Research
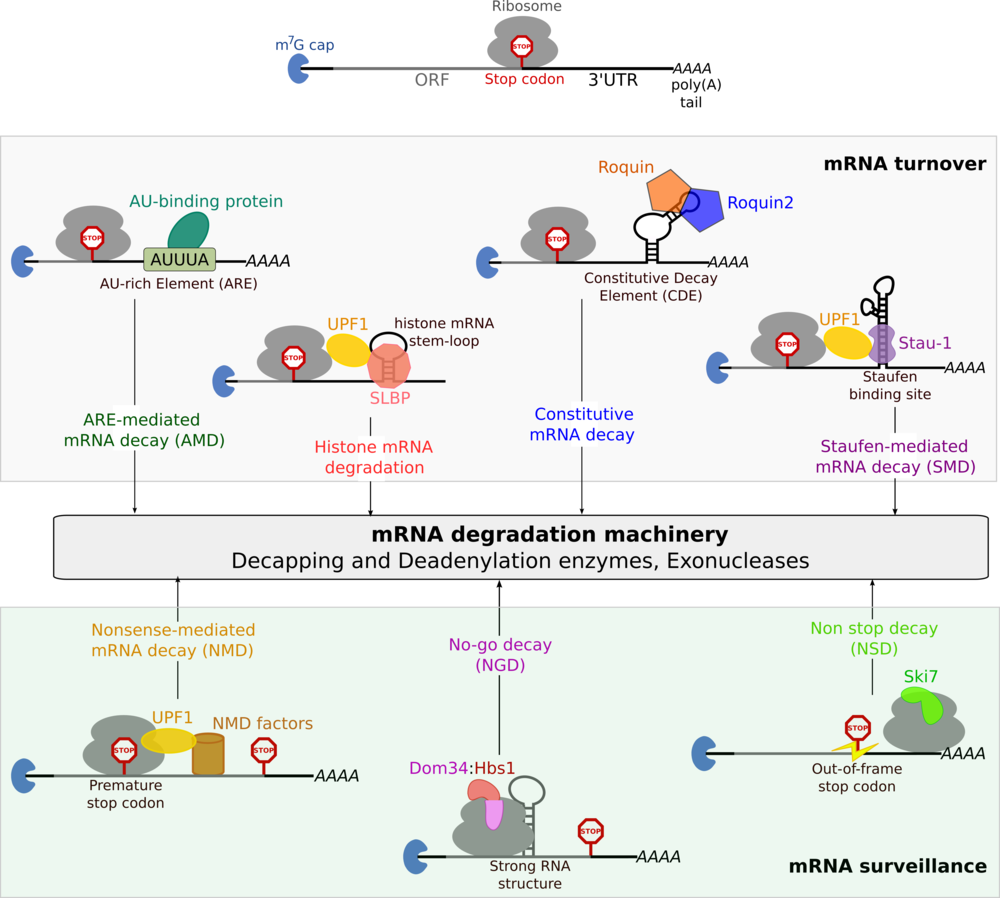
Gene expression in eukaryotes is a complex, multi-step process, subject to stringent regulation at every stage. This regulation can be mediated at the level of DNA (differential transcription) or protein (selective degradation) or at an intermediate step, at the level of mRNA. Post-transcriptional gene regulation occurs at several stages in the life-cycle of an mRNA, such as processing, export, translation and degradation. Degradation of mRNA often serves as a mechanism for achieving rapid changes in gene expression profiles in response to external stimuli. The mRNA transcripts of proto-oncogenes, cytokines and transcription factors are highly labile and are degraded within a few minutes, while transcripts of house-keeping genes have a long half-life. Differential rates of mRNA decay are determined by specific sequence elements, known as cis-acting signals, within the mRNA. Protein factors recognise the cis-acting signals and target the mRNA for decay through a specific pathway (see figure below). There exist several pathways for regulating mRNA levels (mRNA turnover) and for controlling the quality of the mRNA to be translated (mRNA surveillance). All pathways of degradation eventually target the mRNA to the common degradation machinery comprising decapping and deadenylation factors as well as exonucleases (see figure below). It is generally thought that the cis-acting signals and their associated protein factors play a role in targeting the mRNA to the degradation machinery. To date, much work has been done on the architecture and the function of the mRNA degradation machinery. However, an important question in the field of mRNA decay still remains unanswered: how does the cis-acting signal within an mRNA target it to a specific degradation pathway? We are investigating pathways of mRNA decay to determine how the mRNPs connect to the mRNA degradation machinery using a structural biochemistry approach.