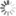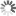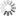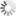New approach to diminish antibiotic resistance developed by research team from Freie Universität Berlin, the Hebrew University of Jerusalem, and ETH Zurich
Pseudomonas aeruginosa unter dem Rasterelektronenmikroskop
Image Credit: Maren Herzog
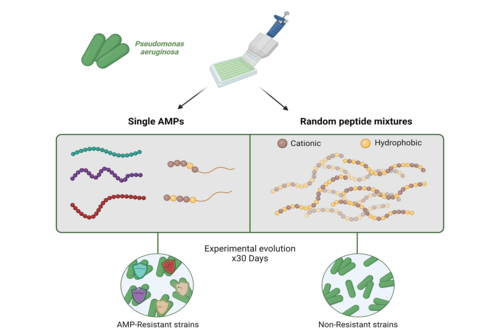
Abbildung zur experimentellen Evolution von Pseudomonas
Image Credit: Bar Manon
News from Jul 04, 2024
Antibiotics are a cornerstone of modern medicine. They are regularly used to treat bacterial infections or as a prophylactic measure during surgery to prevent infection. However, due to the widespread use of antibiotics, many bacteria have grown resistant to them, a development that has dramatic consequences. It is estimated that antibiotic resistant pathogens are responsible for approximately 5 million deaths annually. Even in light of advances in diagnostics and efforts to be more prudent with prescribing antibiotics, new types of drugs will need to be developed in the future to mitigate the impact of increasingly resistant bacteria. One possibility is to use different combinations of antibiotics that make it difficult for bacteria to develop resistance in the first place. A recent study published in PLOS Biology suggests a new possibility that explores how a mixture made up of a million different molecules can also inhibit resistance. The article, “The Evolution of Antimicrobial Peptide Resistance in Pseudomonas aeruginosa Is Severely Constrained by Random Peptide Mixtures,” is available online at https://doi.org/10.1371/journal.pbio.3002692.
Researchers from Freie Universität Berlin, the Hebrew University of Jerusalem, and ETH Zurich worked together to investigate if and how Pseudomonas aeruginosa acquires resistance to new antibiotics. According to the World Health Organization, this bacterium is one of the most prevalent pathogens. The researchers’ approach focused on the use of antimicrobial peptides (AMPs), amino acid chains that can be found in all domains of life, to disrupt the evolution of drug resistance in bacteria. These newly developed random antimicrobial peptide mixtures (RPMs) produce “peptide libraries” that contain a cocktail of a million peptide combinations. One such peptide consists of two amino acids that have been assembled at random. “Over a period of four weeks, which is usually how long treatment lasts for a patient with pneumonia caused by Pseudomonas aeruginosa, we could produce resistance against a wide variety of treatments in our lab experiments. However, we were not able to do so for the random peptide mixture,” says first author of the study Bernardo Antunes, an evolutionary biologist at Freie Universität Berlin. The results of the experiments were further supported by controlled evolution in the lab, genome analyses, and mathematical modeling. “It will still be quite some time before we are ready for practical applications,” says Jens Rolff, one of the principal investigators of the study and biology professor at Freie Universität Berlin. “Still, our current work demonstrates the potential that these combinations have when it comes to reducing antimicrobial resistance.”
Further Information
Publication
Antunes, B., C. Zanchi, P. R. Johnston, B. Maron, C. Witzany, et al. 2024. "The Evolution of Antimicrobial Peptide Resistance in Pseudomonas aeruginosa Is Severely Constrained by Random Peptide Mixtures." PLOS Biology 22, no. 7: e3002692. https://doi.org/10.1371/journal.pbio.3002692.
Contact
Prof. Dr. Jens Rolff, Institute of Biology, Freie Universität Berlin, Tel.: +49 (0)30 385 4893, Email: jens.rolff@fu-berlin.de; Web: https://www.bcp.fu-berlin.de/en/biologie/arbeitsgruppen/zoologie/ag_rolff/index.html